Humans are living ecosystems. Our skin, our lungs and our intestines are all home to complex communities of microorganisms. In fact, the relationship is even deeper, as humans and microbes have adapted to each other in the course of their evolution. Prof. Dr Pierre Stallforth leads a team of researchers in the Cluster of Excellence »Balance of the Microverse«, investigating the natural products that shaped the microbiomes of Neanderthals and modern humans.
By Ute Schönfelder
The primary aim of this paleotechnobiological research is not to analyse the dental or digestive problems experienced by our ancestors. Instead, as Pierre Stallforth and his team examine the human microbiome of the past, their primary interest is finding hitherto unknown natural products that could benefit us today and in the future. »Natural products biosynthesized by microorganisms are an indispensable element of modern medicine,« says Stallforth. »They are the basis for many antibiotics, immunomodulators, and anti-cancer drugs.«
This has instigated an intensive search for as yet unknown sources of natural products, with researchers scouring the oceans, searching deep underground and examining exotic plants and fungi. In addition to exploring new environments and organisms in the search for bioactive molecules, the Jena-based researchers are turning their attention to the distant past.
Molecular biological time travel
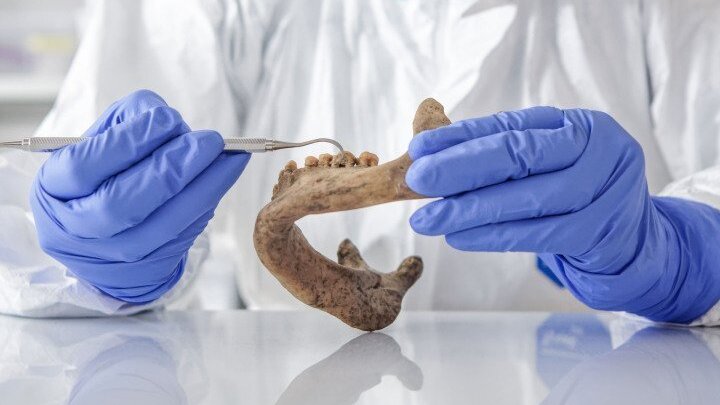
Accessing these natural products hidden in the past means examining the remains of microorganisms that have been preserved for hundreds and even thousands of years. Two sources of human fossils are particularly suitable for this research: coprolites, which are fossilized faeces, and dental tartar. Dental calculus is the only material in the human body that calcifies while we are still alive. The resulting material, known as tartar or calculus, is effectively a mineralized »bacterial graveyard«. The solid framework of calcium phosphate bonds in tartar provides an excellent matrix for the long-term preservation of microbial DNA.
Pre-historic calculus therefore opens the door to reconstructing our ancestors’ microbiome. Nevertheless, the road to identifying pre-historic natural products generated by microbes is still a long one—and relies on a considerable amount of cutting-edge technology. »Firstly, we have to isolate and sequence the DNA samples,« outlines Stallforth. After thousands of years, bacterial genomes are not usually intact or complete. Consequently, the researchers have to reconstitute the genome from numerous parts, which are like pieces of a genetic jigsaw puzzle. Once the Stone Age genetic information has been reconstructed, bioinformatic methods can be applied to search for genes capable of synthesizing bacterial natural products.
Jurassic Park in the lab
Pierre Stallforth and his team have demonstrated the viability of this concept on numerous occasions. In 2023, for example, they examined and compared the tartar of Neanderthals and modern humans in a highly regarded paper published in »Science«. The researchers were able to determine that in addition to an array of microbes that also appear in contemporary samples, oral flora from the Stone Age contain numerous bacteria that no longer exist. »It’s possible that these microbes died out with the Neanderthals. Or perhaps they were displaced by climatic changes or changes in our diet and lifestyle,« says Stallforth.
A calculus sample taken from a roughly 18,500-year-old skeleton of a woman discovered in Spain in 2010 proved particularly fruitful. The researchers discovered the genomes of a variety of the Chlorobium species—a green sulphur bacterium capable of photosynthesis. It is likely that these bacteria originated from the water sources from which people at the time sourced drinking water.
The researchers scoured the reconstructed Chlorobium genomes for biosynthetic genes, which Pierre Stallforth describes as thrilling work. »It was a little like Jurassic Park,« he says. »While these genes provide a blueprint for enzymes, we didn’t know what reactions they would catalyse.« To find out, the researchers implanted the Stone Age genes into modern bacteria—effectively bringing them »back to life«. Stallforth and his team named some of the substances produced by the bacteria »paleofurans«. What the researchers don’t yet know, however, is the functions that these pre-historic natural products introduced in bacteria. »They don’t appear to be antibiotics,« says Stallforth.
Dynamic microbiome
There are a number of potential reasons why the bacteria from the tartar might have produced paleofurans. Pierre Stallforth suspects that they served as signal molecules. For the time being, however, further investigation is required.
Even if it has not yet been possible to attribute a specific function to these paleofurans, this outcome still represents a win for the researchers. »We’ve been able to show, firstly, that it’s fundamentally possible to recreate natural products from extinct microorganisms. Secondly, we’re gaining insights into the chemical composition of a microbiome that no longer exists.«
Stallforth emphasizes that these are significant findings. He explains that he and his team at the Cluster are also analysing the dynamic structure of natural products in microbial communities. »Each substance produced by microorganisms has a function which is not limited to that species alone. Other organisms in the microbiome also detect these substances and, in some cases, chemically modify them to produce new properties or functions.« This is how organisms communicate and alter their environment. »The chemistry of a microbial ecosystem is in a constant state of flux.«
07745 Jena Google Maps site planExternal link