The vast majority of people probably hadn’t ever heard of a coronavirus before 2020. However, the crown-shaped viruses have long been studied by experts. Until a few years ago, they were considered fairly harmless agents that only caused mild cold-like symptoms in humans. But oh how things have changed… Following in the footsteps of SARS and MERS, SARS-CoV-2 is now the third »novel coronavirus« that can trigger a life-threatening disease in people.
Someone who has spent many years dealing with familiar and novel coronaviruses is Prof. Dr. Manja Marz and her team of bioinformaticians at the University of Jena. Manja Marz directs the European Virus Bioinformatics CenterExternal link (EVBC), which was established at the University of Jena in 2017. The centre now has 166 researchers from 28 countries, including both bioinformaticians and virologists, such as Prof. Christian Drosten from the Charité in Berlin.
Corona research tools
»As reflected quite clearly by the current situation, viruses pose a serious threat to human health. That can not only be said of viruses that are yet to be discovered, such as SARS-CoV-2 at the moment, but also viruses that have been known for a long time and constantly re-emerge in different areas of the world«, explains Prof. Marz. She emphasizes the importance of research: »Although bioinformatics is still in its infancy, it enables us to address many basic questions in virology using instruments that are suitable for handling large volumes of data, such as from high-throughput sequencing processes«, says Marz.
Needless to say, the work carried out at the EVBC is currently focused on SARS-CoV-2. »The scientific community has reacted incredibly quickly to the emergence of the novel virus«, says Dr Franziska Hufsky, the scientific coordinator of the EVBC. This is important, because the virus will be difficult to control without a comprehensive understanding of its evolution and pathogenesis. That’s why the EVBC is gathering bioinformatic tools and information for the global research community. On its website, the EVBC lists tools that are specially designed for research into SARS-CoV-2External link. The spectrum ranges from simple virus detection and the sequencing of virus genomes to the distribution and evolution of specific virus strains and tools that can be used to identify suitable targets for drugs.
Franziska Hufsky is a bioinformatician and the scientific coordinator of the European Virus Bioinformatics Centre at the University of Jena. She recently published a review in which she summarizes the most important bioinformatic tools for researching SARS-CoV-2.
Image: Anne Günther (University of Jena)One coronavirus causes a cold – the other triggers COVID-19
Franziska Hufsky, Manja Marz and their team of researchers at the University of Jena recently published a summary External linkof the most important tools that over 50 international researchers have made available to everyone free of charge. Of course, the summary itself can also be accessed free of charge via the EVBC website. »The corona crisis has shown just how well science can work when everything comes together: international, interdisciplinary and cooperative«, says Franziska Hufsky, and the bioinformatician underlines the important role played by institutions like the EVBC: »They’re a melting pot for scientific exchange and act as think tanks and workshops, advancing research developments at a much quicker rate«.
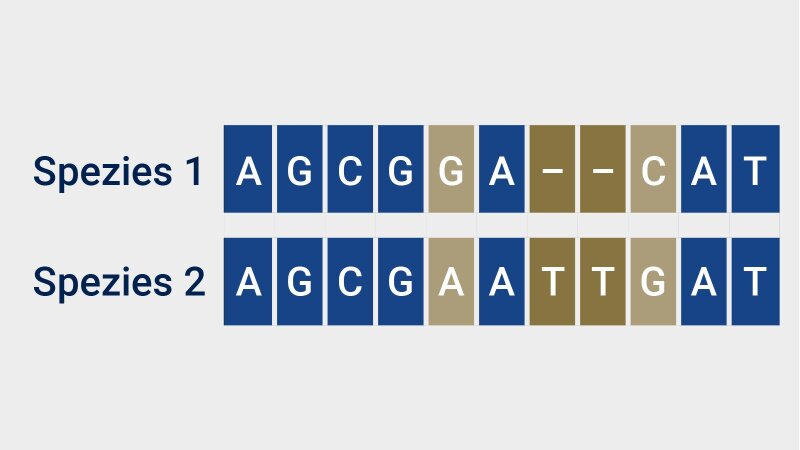
The work carried out by Hufsky and her colleagues is not just limited to the scientific coordination of the EVBC; the bioinformaticians at the University of Jena are also conducting their own virus research and making their results available to others in the community, such as Kevin Lamkiewicz. The doctoral researcher is creating and examining viral alignments: »This enables the comparison of the genome sequences of different species, as identical or very similar sequence segments are arranged on top of one another«, explains Lamkiewicz. By arranging the genomes in this way, we can discover similarities and differences in genomes and, above all, identify sequence segments that might affect the properties of encoded proteins.
For example, the bioinformaticians are currently studying alignments of the novel SARS-CoV-2 coronavirus with human coronavirus 229E (HCoV-229E), a cold virus that has been known for decades. »We’re interested in finding out how HCoV-229E merely causes a harmless cold, while SARS-CoV-2 can shut down the whole world in a matter of weeks«.
Kevin Lamkiewicz, a doctoral candidate in the field of bioinformatics, is investigating the secondary structure of RNA viruses, including SARS-CoV-2.
Image: Anne Günther (University of Jena)Not every difference in genome sequences affects the functionality of virus proteins; in fact, some of the copying errors (mutations) that occur during the viral replication process often go unnoticed (»silent mutations«). »However, some other mutations can prevent a virus from multiplying any further«, explains Lamkiewicz. That’s exactly what he and his colleagues managed to prove in their study of HCoV-229EExternal link: The replication of the virus is drastically reduced when there is a guanine instead of a cytosine in position 47. The researchers now want to identify such areas for SARS-CoV-2, as these points would be promising targets for potential antiviral treatments.
By aligning the genome sequence segments of two species (e.g. viruses), we can superimpose and compare equivalent areas. Identical sequence segments are shown in blue. The pale gold areas show differences that may have resulted from mutations. The gold coloured segment highlights a deletion in species 1.
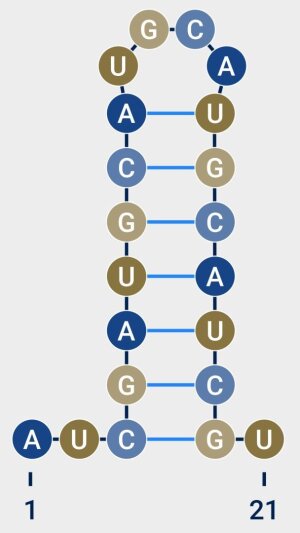
Picture: Kevin Lamkiewicz, bearbeitet von Liana FrankeRNA secondary structure. The blue lines represent interactions between nucleotide bases that are not adjacent and therefore lead to the formation of spatial structures in the RNA.
Picture: Kevin Lamkiewicz, bearbeitet von Liana FrankeThe fact that relatively small changes in genetic information can have enormous effects on virus properties is due to the secondary structure of the genome sequences. The genome sequences of coronaviruses consist of RNA (ribonucleic acid), i.e. single-stranded chains of nucleotides in a row. The individual nucleotides can interact with one another, even if they are not directly adjacent in the chain. This creates structural elements, such as hairpin-shaped paired strands or loops, which can be essential for the replication of the virus. »Changes in the nucleotide sequence can lead to changes in the secondary structure which, in turn, can affect its functionality«, explains Kevin Lamkiewicz.
The bioinformaticians at the University of Jena have created partial alignments for SARS-CoV-2 and all known coronaviruses (including HCoV-229E) for the genome segments that are important for the formation of RNA secondary structures. Their results have been added to the »Rfam« database. The partial alignments can now be accessed by experimental research teams around the world to see whether these structures can be used as starting points for treatments.
Complete virus genome is supplied by mini sequencing device
Needless to say, the genome sequences of viruses can only be studied if the viruses are known in the first place. And that’s exactly what the bioinformaticians are working on at the University of Jena. Since March, they have been working regularly with researchers from the University Hospital to sequence samples from COVID-19 patients as part of the national NFDI4Microbiota External linkconsortium. They’ve been using the MinION system, a small mobile sequencing device that is hardly bigger than a USB stick and can be connected to a laptop or PC (see photo above).
This is an established piece of sequencing technology at the University of Jena. The MinION system is not only conveniently small and easy to use; Sebastian Krautwurst, a doctoral researcher working on the project, explains some of its additional practical advantages over previous sequencing methods: »On the one hand, we can now sequence entire genomes at once, whereas previous methods usually only supplied partial sequences that subsequently had to be pieced together«. On the other hand, the mini sequencer enables the direct analysis of RNA – in the past, it first had to be transferred to DNA.
Sebastian Krautwurst is a doctoral researcher who is sequencing RNA from COVID-19 patient samples.
Image: Anne Günther (University of Jena)When sequencing with the MinION device, individual DNA or RNA strands are pulled through tiny tunnels known as »nanopores«. Changes in the flow of electric current are measured as the individual nucleobases (adenine, guanine, cytosine, thymine or uracil) pass through the pore. The changes are specific for each base, enabling the subsequent determination of their exact sequence.
Sebastian Krautwurst and his colleagues have already sequenced different strains of HCoV-229E and transmissible gastroenteritis virus (TGEV), and they are now turning their attention to the sequencing of SARS-CoV-2 samples.
By Ute Schönfelder